Research
Research Overview
The cell is composed of a highly dynamic proteome that is organized into organelles, structural components, and signaling pathways that encode cellular functions. These components are subject to numerous quality control pathways that ensure their proper organization and remove components that are damaged using autophagy and or the ubiquitin-proteasome system (UPS). At the same time, numerous individual proteins have short halflifes that reflect regulatory signals controlling their degradation by the UPS. These pathways are often disrupted in neurodegenerative diseases, ranging from Parkinson's to Alzheimer's to ALS. Our lab studies these pathways broadly using a variety of approaches, including quantitative proteomics, live cell imaging, and biochemistry, and employs engineered stem cells systems that allow us to make disease relevant models for protein quality control. In addition, we are involved in a major effort to elucidate the organization and structure of the human interactome using interaction proteomics, and have created BioPlex 3.0 containing protein interactions for the majority of human proteins in multiple cell lines.
BioPlex - Human Interactome
The human interactome is composed on thousands of interactions and protein communities. Using a robust interaction proteomics platform, we - together with the Gygi lab - have performed interaction proteomics on more than 10,000 human genes in 2 cell lines, thereby creating BioPlex 3.0 - the largest such network in existence. We are currently expanding this system to additional cell lines to understand how cellular diversity affects the organization of the proteome.
Cullin RING Ubiquitin Ligases
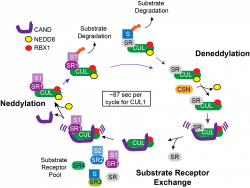
CRLs represent represent the largest collection of E3 ubiquitin ligases and contain a cullin scaffold and one of more than 200 substrate specific adaptors. We have employed various approaches to understand the regulation of CRLs, including by the co-E3 ARIH1, as well as the substrates of individual cullins. Currently, we are examining the role of CRLs and their regulatory apparatus in germ cell development using engineered ES cells together with protocols that allow conversion to endoderm, mesoderm and ectoderm. The goal is to understand the identity of specific CRLs that catalyze specific developmental steps and the targets whose degradation is necessary for specific developmental decisions. This effort is aided by our efforts to develop methods to capture ubiquitin remant-based proteomics.
Organelle quality control and neurodegenerative disease mechanisms
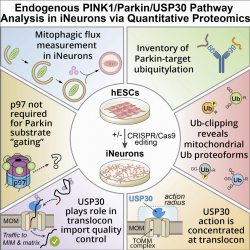
Selective autophagy is a pathway by which organelles, protein aggregates, or in some cases individual proteins are selectively engulfed into a double membrane structure called an autophagosome, which fuses with the lysosome to facilitate degradation and recycling of the damaged material. We study numerous aspects of this pathway and have identified machinery required for signaling, recognizing, and eliminating such organelles. This includes NCOA4 involved in ferritinophagy and TEX264 involved in ER-phagy. In addition, we have worked extensively on the process of mitophagy, which is catalyzed by the PARKIN ubiquitin ligase and the PINK1 protein kinase, two proteins mutated in Parkinson's disease. We are currently working on multiple aspects of ER-phagy, mitophagy, and lysophagy - the process by which damaged lysosomes themselves are degraded. In addition, we are exploring several aspects of the endolysosomal system, including the affects of Lysosomal Storage Disease (LSD) disease genes on lysosomal quality control and have developed a method for isolation of early endosomes for proteomic analysis. In recent work, we have developed a targeted proteomic approach for measuring processing of the Alzheimer's protein APP by beta- and gamma-secretase within both endosomes and lysosomes. Finally, we are exploring the pathways by which protein aggregates linked with neurodegenerative diseases are removed from cells by autophagy and the ubiquitin system.