Publications
DMXL1 promotes recruitment of V1-ATPase to lysosomes upon TRPML1 activation. Lee C, Eldridge MJG, Gonzalez-Lozano MA, Bresnahan T, Niday Z, Del Camino D, Fu T, Paulo JA, Moran MM, Helaine S, Harper JW. Nat Struct Mol Biol. 2025 Jun 17:10.1038/s41594-025-01581-x. doi: 10.1038/s41594-025-01581-x. Epub ahead of print. PMID: 40527988; PMCID: PMC12246789. |
|
EndoMAP.v1 charts the structural landscape of human early endosome complexes. Gonzalez-Lozano MA, Schmid EW, Miguel Whelan E, Jiang Y, Paulo JA, Walter JC, Harper JW. Nature. 2025 May 28. doi: 10.1038/s41586-025-09059-y. Epub ahead of print. PMID: 40437099. |
|
Global cellular proteo-lipidomic profiling of diverse lysosomal storage disease mutants using nMOST. Kraus F, He Y, Swarup S, Overmyer KA, Jiang Y, Brenner J, Capitanio C, Bieber A, Jen A, Nightingale NM, Anderson BJ, Lee C, Paulo JA, Smith IR, Plitzko JM, Gygi SP, Schulman BA, Wilfling F, Coon JJ, Harper JW. bioRxiv [Preprint]. 2024 Oct 20:2024.03.26.586828. doi: 10.1101/2024.03.26.586828. PMID: 38585873; PMCID: PMC10996675. |

|
Global cellular proteo-lipidomic profiling of diverse lysosomal storage disease mutants using nMOST. Kraus F, He Y, Swarup S, Overmyer KA, Jiang Y, Brenner J, Capitanio C, Bieber A, Jen A, Nightingale NM, Anderson BJ, Lee C, Paulo JA, Smith IR, Plitzko JM, Gygi SP, Schulman BA, Wilfling F, Coon JJ, Harper JW. Sci Adv. 2025 Jan 24;11(4):eadu5787. doi: 10.1126/sciadv.adu5787. Epub 2025 Jan 22. PMID: 39841834. |
|
Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases. Scott DC, Chittori S, Purser N, King MT, Maiwald SA, Churion K, Nourse A, Lee C, Paulo JA, Miller DJ, Elledge SJ, Harper JW, Kleiger G, Schulman BA. Nat Commun. 2024 Nov 15;15(1):9899. doi: 10.1038/s41467-024-54126-z. PMID: 39548056; PMCID: PMC11568203. |

|
Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2. Scott DC, Dharuman S, Griffith E, Chai SC, Ronnebaum J, King MT, Tangallapally R, Lee C, Gee CT, Yang L, Li Y, Loudon VC, Lee HW, Ochoada J, Miller DJ, Jayasinghe T, Paulo JA, Elledge SJ, Harper JW, Chen T, Lee RE, Schulman BA. Nat Commun. 2024 Oct 12;15(1):8829. doi: 10.1038/s41467-024-52966-3. PMID: 39396041; PMCID: PMC11470957. |

|
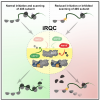
IRGQ-mediated autophagy in MHC class I quality control promotes tumor immune evasion. Herhaus L, Gestal-Mato U, Eapen VV, Mačinković I, Bailey HJ, Prieto-Garcia C, Misra M, Jacomin AC, Ammanath AV, Bagarić I, Michaelis J, Vollrath J, Bhaskara RM, Bündgen G, Covarrubias-Pinto A, Husnjak K, Zöller J, Gikandi A, Ribičić S, Bopp T, van der Heden van Noort GJ, Langer JD, Weigert A, Harper JW, Mancias JD, Dikic I. Cell. 2024 Dec 12;187(25):7285-7302.e29. doi: 10.1016/j.cell.2024.09.048. Epub 2024 Oct 30. PMID: 39481378. |

|
Endo-IP and lyso-IP toolkit for endolysosomal profiling of human-induced neurons. Hundley FV, Gonzalez-Lozano MA, Gottschalk LM, Cook ANK, Zhang J, Paulo JA, Harper JW. Proc Natl Acad Sci U S A. 2024 Dec 24;121(52):e2419079121. doi: 10.1073/pnas.2419079121. Epub 2024 Dec 5. PMID: 39636867; PMCID: PMC11670117. |

|
Hickey KL, Panov A, Whelan EM, Schäfer T, Mizrak A, Kopito RR, Baumeister W, Fernández-Busnadiego R, Harper JW. Proc Natl Acad Sci U S A. 2024 Dec 10;121(50):e2417390121. doi: 10.1073/pnas.2417390121. Epub 2024 Dec 5. PMID: 39636856; PMCID: PMC11648907. |

|
Spatiotemporal proteomic profiling of cellular responses to NLRP3 agonists. Hollingsworth LR, Veeraraghavan P, Paulo JA, Harper JW. bioRxiv [Preprint]. 2024 Apr 20:2024.04.19.590338. doi: 10.1101/2024.04.19.590338. PMID: 38659763; PMCID: PMC11042255. |
|
Visualizing chaperone-mediated multistep assembly of the human 20S proteasome. Adolf F, Du J, Goodall EA, Walsh RM Jr, Rawson S, von Gronau S, Harper JW, Hanna J, Schulman BA. Nat Struct Mol Biol. 2024 Apr 10. doi: 10.1038/s41594-024-01268-9. Epub ahead of print. PMID: 38600324. |
|
Combinatorial selective ER-phagy remodels the ER during neurogenesis. Hoyer MJ, Capitanio C, Smith IR, Paoli JC, Bieber A, Jiang Y, Paulo JA, Gonzalez-Lozano MA, Baumeister W, Wilfling F, Schulman BA, Harper JW. Nat Cell Biol. 2024 Mar;26(3):378-392. doi: 10.1038/s41556-024-01356-4. Epub 2024 Mar 1. PMID: 38429475; PMCID: PMC10940164. |
|
UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER DaRosa PA, Penchev I, Gumbin SC, Scavone F, Wąchalska M, Paulo JA, Ordureau A, Peter JJ, Kulathu Y, Harper JW, Becker T, Beckmann R, Kopito RR. Nature. 2024 Mar;627(8003):445-452. doi: 10.1038/s41586-024-07073-0. Epub 2024 Feb 21. PMID: 38383785. |
|
Goldsmith J, Ordureau A, Harper JW, Holzbaur ELF.Neuron. 2024 Feb 7;112(3):520. doi: 10.1016/j.neuron.2024.01.011.PMID: 38330901 |
|
Watts ME, Giadone RM, Ordureau A, Holton KM, Harper JW, Rubin LL.Front Cell Neurosci. 2024 Jan 19;17:1327361. doi: 10.3389/fncel.2023.1327361. eCollection 2023.PMID: 38314348 |
|
Springstein BL, Paulo JA, Park H, Henry K, Fleming E, Feder Z, Harper JW, Hochschild A.Proc Natl Acad Sci U S A. 2024 Feb 6;121(6):e2317453121. doi: 10.1073/pnas.2317453121. Epub 2024 Jan 30. |
|
Panov A, Harper JW. 2023 Oct 31;3(1):vbad157. doi: 10.1093/bioadv/vbad157. PMID: 37953912; PMCID: PMC10636703. |
|
Proteome census upon nutrient stress reveals Golgiphagy membrane receptors. |
|
ER membrane curvature and ubiquitin as drivers of ER-phagy. Hoyer MJ, Harper JW. Dev Cell. 2023 Jul 24;58(14):1219-1220. doi: 10.1016/j.devcel.2023.06.008. PMID: 37490852. |
|
Proteome census upon nutrient stress reveals Golgiphagy membrane receptors. Hickey KL, Swarup S, Smith IR, Paoli JC, Miguel Whelan E, Paulo JA, Harper JW. Nature. 2023 Sep 27. doi: 10.1038/s41586-023-06657-6. Epub ahead of print. PMID: 37757899. |
|
Mechanisms Controlling Selective Elimination of Damaged Lysosomes. Hoyer MJ, Swarup S, Harper JW.Curr Opin Physiol. 2022 Oct;29:100590. doi: 10.1016/j.cophys.2022.100590. Epub 2022 Aug 30.PMID: 36713230 |
|
Substitution of PINK1 Gly411 modulates substrate receptivity and turnover. Fiesel FC, Fričová D, Hayes CS, Coban MA, Hudec R, Bredenberg JM, Broadway BJ, Markham BN, Yan T, Boneski PK, Fiorino G, Watzlawik JO, Hou X, McCarty AM, Lewis-Tuffin LJ, Zhong J, Madden BJ, Ordureau A, An H, Puschmann A, Wszolek ZK, Ross OA, Harper JW, Caulfield TR, Springer W.Autophagy. 2022 Dec 5:1-22. doi: 10.1080/15548627.2022.2151294. Online ahead of print.PMID: 36469690 |
|
|
Park H, Hundley FV, Yu Q, Overmyer KA, Brademan DR, Serrano L, Paulo JA, Paoli JC, Swarup S, Coon JJ, Gygi SP, Harper JW. Nat Commun. 2022 Oct 16;13(1):6112. doi: 10.1038/s41467-022-33881-x.PMID: 36245040 |
|
Deficiency of the frontotemporal dementia gene GRN results in gangliosidosis. Boland S, Swarup S, Ambaw YA, Malia PC, Richards RC, Fischer AW, Singh S, Aggarwal G, Spina S, Nana AL, Grinberg LT, Seeley WW, Surma MA, Klose C, Paulo JA, Nguyen AD, Harper JW, Walther TC, Farese RV Jr.Nat Commun. 2022 Oct 7;13(1):5924. doi: 10.1038/s41467-022-33500-9.PMID: 36207292 |
|
Welsh KA, Bolhuis DL, Nederstigt AE, Boyer J, Temple BRS, Bonacci T, Gu L, Ordureau A, Harper JW, Steimel JP, Zhang Q, Emanuele MJ, Harrison JS, Brown NG.EMBO J. 2022 Feb 1;41(3):e108823. doi: 10.15252/embj.2021108823. Epub 2021 Dec 23.PMID: 34942047 |
|
Mechanisms underlying ubiquitin-driven selective mitochondrial and bacterial autophagy. Goodall EA, Kraus F, Harper JW.Mol Cell. 2022 Apr 21;82(8):1501-1513. doi: 10.1016/j.molcel.2022.03.012. Epub 2022 Mar 31.PMID: 35364016 |
|
|
Goldsmith J, Ordureau A, Harper JW, Holzbaur ELF.Neuron. 2022 Mar 16;110(6):967-976.e8. doi: 10.1016/j.neuron.2021.12.029. Epub 2022 Jan 19.PMID: 35051374
|
|
|
Antico O, Ordureau A, Stevens M, Singh F, Nirujogi RS, Gierlinski M, Barini E, Rickwood ML, Prescott A, Toth R, Ganley IG, Harper JW, Muqit MMK.Sci Adv. 2021 Nov 12;7(46):eabj0722. doi: 10.1126/sciadv.abj0722. Epub 2021 Nov 12.PMID: 34767452
|
|
|
Molecular Cell. 2021 Oct 15:S1097-2765(21)00800-5. doi: 10.1016/j.molcel.2021.10.001. Online ahead of print.
|

|
Sebastian Boland*, Sharan Swarup*, Yohannes A Ambaw, Ruth C Richards, Alexander W Fischer, Shubham Singh, Geetika Aggarwal, Salvatore Spina, Alissa L Nana, Lea T Grinberg, William W Seeley, Michal A Surma, Christian Klose, Joao A Paulo, Andrew D Nguyen, J. Wade Harper, Tobias C Walther, Robert V Farese Jr. (2021) Progranulin deficiency results in reduced bis(monoacylglycero)phosphate (BMP) levels and gangliosidosis. https://www.biorxiv.org/content/10.1101/2021.09.30.461806v1. *, equal contribution. |
|
Sharan's work with Sebastian Boland in the Walther/Farese lab has been posted on BioRxiv. This work, which uses Lyso-IP and lipidomics reveales that Progranulin deficiency results in reduced bis(monoacylglycero)phosphate (BMP) levels and gangliosidosis. Very cool study!
|
|
Eapen, V., Swarup, S., Hoyer, M. J., Paulo, J. A. & Harper, J. W. Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy. Elife 10, e72328., doi: 10.7554/eLife.72328. (2021). |
|
iRQC, a surveillance pathway for 40S ribosomal quality control during mRNA translation initiation Garshott DM, An H, Sundaramoorthy E, Leonard M, Vicary A, Harper JW, Bennett EJ. (2021), Cell Reports (https://www.cell.com/cell-reports/pdf/S2211-1247(21)01085-8.pdf) |
|
ORF10-Cullin-2-Zyg11B complex is not required for SARS-CoV-2 infection Mena EL, Donahue CJ, Vaites LP, Li J, Rona G, O'Leary C, Lignitto L, Miwatani-Minter B, Paulo JA, Dhabaria A, Ueberheide B, Gygi SP, Pagano M, Harper JW, Davey RA, Elledge SJ. Proc Natl Acad Sci U S A. 2021 Apr 27;118(17):e2023157118. doi: 10.1073/pnas.2023157118. PMID: 33827988 |
|
Host ubiquitin protein tags lipid to fight bacteria Schulman BA, Harper JW. Nature. 2021 Jun;594(7861):28-29. doi: 10.1038/d41586-021-01267-6. PMID: 34012123 No abstract available. |
|
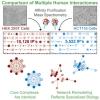
Dual proteome-scale networks reveal cell-specific remodeling of the human interactome Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, Gebreab F, Gygi MP, Thornock A, Zarraga G, Tam S, Szpyt J, Gassaway BM, Panov A, Parzen H, Fu S, Golbazi A, Maenpaa E, Stricker K, Guha Thakurta S, Zhang T, Rad R, Pan J, Nusinow DP, Paulo JA, Schweppe DK, Vaites LP, Harper JW*, Gygi SP*. Cell. 2021 May 27;184(11):3022-3040.e28. doi: 10.1016/j.cell.2021.04.011. Epub 2021 May 6. PMID: 33961781. *, Co-corresponding. |
|
|
Wrighton PJ, Shwartz A, Heo JM, Quenzer ED, LaBella KA, Harper JW, Goessling W. J Cell Sci. 2021 Feb 22;134(4):jcs256255. doi: 10.1242/jcs.256255. PMID: 33536245 |
|
Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-box hypothesis Harper JW, Schulman BA. Annu Rev Biochem. 2021 Jun 20;90:403-429. doi: 10.1146/annurev-biochem-090120-013613. Epub 2021 Apr 6. PMID: 33823649 |
|
|
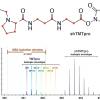
Ordureau A, Yu Q, Bomgarden RD, Rogers JC, Harper JW, Gygi SP, Paulo JA. J Proteome Res. 2021 May 7;20(5):3009-3013. doi: 10.1021/acs.jproteome.0c01056. Epub 2021 Mar 10. PMID: 33689365 |
|
The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. McKenna MJ, Sim SI, Ordureau A, Wei L, Harper JW, Shao S, Park E. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science. 2020 Sep 25;369(6511):eabc5809. doi: 10.1126/science.abc5809. PMID: 32973005. |

|
EDF1 coordinates cellular responses to ribosome collisions. Sinha NK, Ordureau A, Best K, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M, Paulo JA, Harper JW, Bennett EJ, Beckmann R, Green R. EDF1 coordinates cellular responses to ribosome collisions. Elife. 2020 Aug 3;9:e58828. doi: 10.7554/eLife.58828. PMID: 32744497; PMCID: PMC7486125. |
|
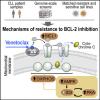
Mitochondrial Reprogramming Underlies Resistance to BCL-2 Inhibition in Lymphoid Malignancies. Guièze R, Liu VM, Rosebrock D, Jourdain AA, Hernández-Sánchez M, Martinez Zurita A, Sun J, Ten Hacken E, Baranowski K, Thompson PA, Heo JM, Cartun Z, Aygün O, Iorgulescu JB, Zhang W, Notarangelo G, Livitz D, Li S, Davids MS, Biran A, Fernandes SM, Brown JR, Lako A, Ciantra ZB, Lawlor MA, Keskin DB, Udeshi ND, Wierda WG, Livak KJ, Letai AG, Neuberg D, Harper JW, Carr SA, Piccioni F, Ott CJ, Leshchiner I, Johannessen CM, Doench J, Mootha VK, Getz G, Wu CJ. Cancer Cell. 2019 Oct 14;36(4):369-384.e13. doi: 10.1016/j.ccell.2019.08.005. Epub 2019 Sep 19. |
|
|
Heo JM, Harper NJ, Paulo JA, Li M, Xu Q, Coughlin M, Elledge SJ, Harper JW. Science Advances. 2019 Nov 6;5(11):eaay4624. doi: 10.1126/sciadv.aay4624. eCollection 2019 Nov. |
|
|
Ahfeldt T, Ordureau A, Bell C, Sarrafha L, Sun C, Piccinotti S, Grass T, Parfitt GM, Paulo JA, Yanagawa F, Uozumi T, Kiyota Y, Harper JW, Rubin LL. Stem Cell Reports. 2020 Jan 14;14(1):75-90. doi: 10.1016/j.stemcr.2019.12.005. Epub 2020 Jan 2. |
|
|
Ordureau A, Paulo JA, Zhang J, An H, Swatek KN, Cannon JR, Wan Q, Komander D, Harper JW. Molecular Cell. 2020 Mar 5;77(5):1124-1142.e10. doi: 10.1016/j.molcel.2019.11.013. |

|
|
Petit CS, Lee JJ, Boland S, Swarup S, Christiano R, Lai ZW, Mejhert N, Elliott SD, McFall D, Haque S, Huang EJ, Bronson RT, Harper JW, Farese RV Jr, Walther TC. Proc Natl Acad Sci U S A. 2020 May 12;117(19):10565-10574. doi: 10.1073/pnas.1913956117. Epub 2020 Apr 28. |
|
Systematic quantitative analysis of ribosome inventory during nutrient stress. An H, Ordureau A, Körner M, Paulo JA, Harper JW. Nature. 2020 Jul;583(7815):303-309. doi: 10.1038/s41586-020-2446-y. Epub 2020 Jul 1. |
|
| Santana-Codina N, Gableske S, Rey MQD, Małachowska B, Jedrychowski MP, Biancur DE, Schmidt PJ, Fleming MD, Fendler W, Harper JW, Kimmelman AC, Mancias JD. NCOA4 maintains murine erythropoiesis via cell autonomous and non-autonomous mechanisms. Haematologica. 2019 Jul;104(7):1342-1354. | |
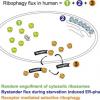
An H, Harper JW. Ribosome Abundance Control Via the Ubiquitin-Proteasome System and Autophagy. J Mol Biol. 2019 Jun 11. pii: S0022-2836(19)30353-5. |
|
| Brennan CM, Vaites LP, Wells JN, Santaguida S, Paulo JA, Storchova Z, Harper JW, Marsh JA, Amon A. Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev. 2019 Aug 1;33(15-16):1031-1047. | |
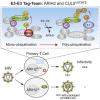
Hüttenhain R, Xu J, Burton LA, Gordon DE, Hultquist JF, Johnson JR, Satkamp L, Hiatt J, Rhee DY, Baek K, Crosby DC, Frankel AD, Marson A, Harper JW, Alpi AF, Schulman BA, Gross JD, Krogan NJ. ARIH2 Is a Vif-Dependent Regulator of CUL5-Mediated APOBEC3G Degradation in HIV Infection. Cell Host Microbe. 2019 Jul 10;26(1):86-99.e7. |
|
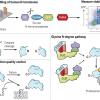
Timms RT, Zhang Z, Rhee DY, Harper JW, Koren I, Elledge SJ. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science. 2019 Jul 5;365(6448). |
|
| Kendrick AA, Dickey AM, Redwine WB, Tran PT, Vaites LP, Dzieciatkowska M, Harper JW, Reck-Peterson SL. Hook3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein-1 and KIF1C. J Cell Biol. 2019 Sep 2;218(9):2982-3001. | |
| Gottlieb CD, Thompson ACS, Ordureau A, Harper JW, Kopito RR. Acute unfolding of a single protein immediately stimulates recruitment of ubiquitin protein ligase E3C (UBE3C) to 26S proteasomes. J Biol Chem. 2019 Aug 2. pii: jbc.RA119.009654. | |
| Jacoupy M, Hamon-Keromen E, Ordureau A, Erpapazoglou Z, Coge F, Corvol JC, Nosjean O, Mannoury la Cour C, Millan MJ, Boutin JA, Harper JW, Brice A, Guedin D, Gautier CA, Corti O. The PINK1 kinase-driven ubiquitin ligase Parkin promotes mitochondrial protein import through the presequence pathway in living cells. Sci Rep. 2019. 9:11829. | |
| Byrum AK, Carvajal-Maldonado D, Mudge MC, Valle-Garcia D, Majid MC, Patel R, Sowa ME, Gygi SP, Harper JW, Shi Y, Vindigni A, Mosammaparast N. Mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function. J Cell Biol. 2019 Feb 4;218(2):422-432. | |
| Kühnle S, Martínez-Noël G, Leclere F, Hayes SD, Harper JW, Howley PM. Angelman syndrome-associated point mutations in the Zn(2+)-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. J Biol Chem. 2018 Nov 23;293(47):18387-18399. | |
Neurohr GE, Terry RL, Lengefeld J, Bonney M, Brittingham GP, Moretto F, Miettinen TP, Vaites LP, Soares LM, Paulo JA, Harper JW, Buratowski S, Manalis S, van Werven FJ, Holt LJ, Amon A. Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence. Cell. 2019 Feb 21;176(5):1083-1097 |

|
Piccolis M, Bond LM, Kampmann M, Pulimeno P, Chitraju C, Jayson CBK, Vaites LP, Boland S, Lai ZW, Gabriel KR, Elliott SD, Paulo JA, Harper JW, Weissman JS, Walther TC, Farese RV Jr. Probing the Global Cellular Responses to Lipotoxicity Caused by Saturated Fatty Acids. Molecular Cell. 2019 Apr 4;74(1):32-44. |

|
| An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Molecular Cell. 2019 Apr 11. pii: S1097-2765(19)30258-8. doi: 10.1016/j.molcel.2019.03.034. [Epub ahead of print] |
|
| Heo JM, Ordureau A, Swarup S, Paulo JA, Shen K, Sabatini DM, Harper JW. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Science Advances. 2018 Nov 21;4(11):eaav0443. |
|
Ordureau A, Paulo JA, Zhang W, Ahfeldt T, Zhang J, Cohn EF, Hou Z, Heo JM, Rubin LL, Sidhu SS, Gygi SP, Harper JW.Dynamics of PARKIN-Dependent Mitochondrial Ubiquitylation in Induced Neurons and Model Systems Revealed by Digital Snapshot Proteomics.Mol Cell. 2018 Apr 19;70(2):211-227.e8. doi: 10.1016/j.molcel.2018.03.012. Epub 2018 Apr 12.PMID:29656925 |
|
Pontano Vaites L, Harper JW.Protein aggregates caught stalling.Nature. 2018 Mar 22;555(7697):449-451. doi: 10.1038/d41586-018-03000-2. No abstract available.PMID:29565390 |

|
Harper JW, Ordureau A, Heo JM.Building and decoding ubiquitin chains for mitophagy.Nat Rev Mol Cell Biol. 2018 Jan 23;19(2):93-108. doi: 10.1038/nrm.2017.129. Review.PMID:29358684 |
|
| An H, Harper JW. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2017 Dec 11. doi:10.1038/s41556-017-0007-x. [Epub ahead of print] |
|
Pontano Vaites L, Paulo JA, Huttlin EL, Harper JW. Systematic analysis of human cells lacking ATG8 proteins uncovers roles for GABARAPs and the CCZ1/MON1 regulator C18orf8/RMC1 in macro and selective autophagic flux. Mol Cell Biol. 2017 Oct 16. doi: 10.1128/MCB.00392-17. PubMed PMID: 29038162. |
|
Scott DC, Hammill JT, Min J, Rhee DY, Connelly M, Sviderskiy VO, Bhasin D, Chen Y, Ong SS, Chai SC, Goktug AN, Huang G, Monda JK, Low J, Kim HS, Paulo JA, Cannon JR, Shelat AA, Chen T, Kelsall IR, Alpi AF, Pagala V, Wang X, Peng J, Singh B, Harper JW, Schulman BA, Guy RK. (2017) Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat Chem Biol. 13(8):850-857. |

|
| Biancur DE, Paulo JA, Małachowska B, Quiles Del Rey M, Sousa CM, Wang X, Sohn ASW, Chu GC, Gygi SP, Harper JW, Fendler W, Mancias JD, Kimmelman AC. (2017) Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat Commun. doi: 10.1038/ncomms15965. |

|
| Liu L, Michowski W, Inuzuka H, Shimizu K, Nihira NT, Chick JM, Li N, Geng Y, Meng AY, Ordureau A, Kołodziejczyk A, Ligon KL, Bronson RT, Polyak K, Harper JW, Gygi SP, Wei W, Sicinski P. (2017) G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat Cell Biol. 19(3):177-188. |

|
| Mohideen F, Paulo JA, Ordureau A, Gygi SP, Harper JW. (2017) Quantitative Phospho-proteomic Analysis of TNFα/NFκB Signaling Reveals a Role for RIPK1 Phosphorylation in Suppressing Necrotic Cell Death. Mol Cell Proteomics. 16(7):1200-1216. |

|
| Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, Szpyt J, Tam S, Zarraga G, Pontano-Vaites L, Swarup S, White AE, Schweppe DK, Rad R, Erickson BK, Obar RA, Guruharsha KG, Li K, Artavanis-Tsakonas S, Gygi SP, Harper JW. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature. 25;545(7655):505-509. |

|
Yang W, Nagasawa K, Münch C, Xu Y, Satterstrom K, Jeong S, Hayes SD, Jedrychowski MP, Vyas FS, Zaganjor E, Guarani V, Ringel AE, Gygi SP, Harper JW, Haigis MC. (2016) Mitochondrial Sirtuin Network Reveals Dynamic SIRT3-Dependent Deacetylation in Response to Membrane Depolarization. Cell. 167(4):985-1000. |

|
| Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu KP, Zhang W, Yu S, Mercredi PY, Harrison JS, Davidson IF, Qiao R, Lu Y, Dube P, Brunner MR, Grace CR, Miller DJ, Haselbach D, Jarvis MA, Yamaguchi M, Yanishevski D, Petzold G, Sidhu SS, Kuhlman B, Kirschner MW, Harper JW, Peters JM, Stark H, Schulman BA. (2016) Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C. Cell. 165(6):1440-53. |
|
| Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P. (2016) Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-Dependent Regulation of PI4P. Cell. 166(2):408-23. |
|
| Harper JW, Bennett EJ. (2016) Proteome complexity and the forces that drive proteome imbalance. Nature. 537(7620):328-38. |
|
| Rose CM, Isasa M, Ordureau A, Prado MA, Beausoleil SA, Jedrychowski MP, Finley DJ, Harper JW, Gygi SP. (2016) Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Systems. pii: S2405-4712(16)30264-2. |

|
| Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, Schulman BA. (2016) Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell. 166(5):1198-1214. |

|
| Guarani V, Jardel C, Chrétien D, Lombès A, Bénit P, Labasse C, Lacène E, Bourillon A, Imbard A, Benoist JF, Dorboz I, Gilleron M, Goetzman ES, Gaignard P, Slama A, Elmaleh-Bergès M, Romero NB, Rustin P, Ogier de Baulny H, Paulo JA, Harper JW, Schiff M. (2016) QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease. Elife. doi: 10.7554/eLife.17163. |

|
Nadarajan S, Mohideen F, Tzur YB, Ferrandiz N, Crawley O, Montoya A, Faull P, Snijders AP, Cutillas PR, Jambhekar A, Blower MD, Martinez-Perez E, Harper JW, Colaiacovo MP. (2016) The MAP kinase pathway coordinates crossover designation with disassembly of synaptonemal complex proteins during meiosis. Elife doi: 10.7554/eLife.12039. |

|
Bui DA, Lee W, White AE, Harper JW, Schackmann RC, Overholtzer M, Selfors LM, Brugge JS. (2016) Cytokinesis involves a nontranscriptional function of the Hippo pathway effector YAP. Sci Signal doi:10.1126/scisignal.aaa9227. |

|
Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, Hu J, Persaud A, Mukherjee M, Li N, Doye A, Walker JR, Sheng Y, Hao Z, Li Y, Brown KR, Lemichez E, Chen J, Tong Y, Harper JW, Moffat J, Rotin D, Schulman BA, Sidhu SS. (2016) System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Molecular Cell 62:121-136. |

|
Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. (2016) The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165:153-164. |

|
Feng Y, Backues SK, Baba M, Heo JM, Harper JW, Klionsky DJ. (2016) Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 12:648-658. |

|
| Raman M, Sergeev M, Garnaas M, Lydeard JR, Huttlin EL, Goessling W, Shah JV, Harper JW. (2015) Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nature Cell Biology 17:1356-1369. |

|
| Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. (2015) The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Molecular Cell 60:7-20. |
|
| Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. (2015) A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Reports 11:1486-1500. |
|
| Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. (2015) Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron 87:371-881. |

|
| Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162:425-440. |

|
Guarani V, McNeill EM, Paulo JA, Huttlin EL, Fröhlich F, Gygi SP, Van Vactor D, Harper JW. (2015) QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. Elife doi: 10.7554/eLife.06265. |
|
Ordureau A, Münch C, Harper JW. (2015) Quantifying Ubiquitin Signaling. Molecular Cell 58:660-676. |
|
Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA*, Harper JW*. (2015) Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A. 112:6637-6642. *, co-corresponding authors |
|
Ordureau A, Sarraf SA, Duda DM, Heo J-M, Jedrykowski MP, Sviderskiy V, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, Wells JA, Gygi SP, Schulman BA, and Harper JW (2014) Quantitative proteomics reveal a feed-forward mechanism for mitochondrial PARKIN translocation and UB chain synthesis. Molecular Cell, 56: 360–375. |

|
Ikeuchi Y, Dadakhujaev S, Chandhoke AS, Huynh MA, Oldenborg A, Ikeuchi M, Deng L, Bennett EJ, Harper JW, Bonni A, Bonni S. (2014) TIF1γ Protein Regulates Epithelial-Mesenchymal Transition by Operating as a Small Ubiquitin-like Modifier (SUMO) E3 Ligase for the Transcriptional Regulator SnoN1. J Biol Chem. 289:25067-25078. |
|
Vaites LL, Harper JW. (2014) Spindle assembly factor protection. Molecular Cell 53:165-166. |
|
Ordureau A, Harper JW. (2014) Cell biology: balancing act. Nature 510:347-348. News and Views |
|
Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. (2014) Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157:1671-1684. |
|
Fischer ES, Böhm K, Lydeard JR, Yang H, Stadler MB, Cavadini S, Nagel J, Serluca F, Acker V, Lingaraju GM, Tichkule RB, Schebesta M, Forrester WC, Schirle M, Hassiepen U, Ottl J, Hild M, Beckwith RE, Harper JW, Jenkins JL, Thomä NH. (2014) Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512:49-53. |
|
Mancias JD, Wang X, Gygi SP, Harper JW*, and Kimmelman AC*. (2014) Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature, 509:105-109. (image: Eric D. Smith and Joseph D. Mancias) |

|
Guarani, V., Paulo, J., Zhai, B., Huttlin, E.L., Gygi, S.P., and Harper, J.W. (2014) TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial Complex I assembly factor through association with the MCIA complex. Molecular and Cellular Biology, 34:847-861. |
|
| Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, Dango S, Luo Y, Hempel K, Sowa ME, Gygi SP, Steen H, Harper JW, Yankner B, Shi Y. (2013) The histone demethylase LSD1/KDM1A promotes the DNA damage response. J Cell Biol. 203:457-470. | |
Mejia LA, Litterman N, Ikeuchi Y, de la Torre-Ubieta L, Bennett EJ, Zhang C, Harper JW, Bonni A. (2013) A Novel Hap1-Tsc1 Interaction Regulates Neuronal mTORC1 Signaling and Morphogenesis in the Brain. J Neurosci. 33:18015-21. |
|
Lydeard JR, Schulman BA, Harper JW. (2013) Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. Nov 15. doi: 10.1038/embor.2013.173. |
|
Tan, M., Lim, S., Bennett, E.J., Shi, Y., and Harper, J.W. (2013) Parallel SCF Adaptor Capture Proteomics Reveals a Role for SCFFBXL17 in NRF2 Activation via BACH1 Repressor Turnover. Molecular Cell, 52, 9-24. |

|
Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Harper JW, andBaehrecke EH. (2013) Uba1 functions in Atg7- and Atg3-independent autophagy.Nature Cell Biology, 15:1067-78. |
|
Zhang C, Mejia LA, Valnegri P, Bennett EJ, Anckar J, Jahani-Asl A, Gallardo G, Ikeuchi Y, Yamada T, Rudnicki M, Harper JW, and Bonni A. (2013) The X-linked Intellectual Disability Protein PHF6 Associates with the PAF1 Complex and Regulates Neuronal Migration in the Mammalian Brain. Neuron 78:986-993 |
|
Sarraf S, Raman M, Guarani-Pereria V, Sowa ME, Huttlin EL, Gygi SP, and Harper JW. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496:372-376. |
|
Martínez-Noël G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM. (2012) Identification and Proteomic Analysis of Distinct UBE3A/E6AP Protein Complexes. Mol Cell Biol 32:3095-3106. |
|
Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I (2012) Rab GAPs in Autophagy: Regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifieers. Mol Cell Biol. 32: 1733-1744. |
|
Tan MJ, White EA, Sowa ME, Harper JW, Aster JC, Howley PM (2012) Cutaneous beta- human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc Natl Acad Sci USA, 109:E1473-80. |
|
Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW and Kopito RR. (2011) Defining human ERAD networks through an integrative mapping strategy. Nature Cell Biology, 14; 93-105 |
|
Kim W, Bennett EJ, Huttlin EL, Guo A., Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW*, and Gygi SP*. (2010) Systematic and quantitative analysis of the ubiquitin modified proteome. Molecular Cell 44:325-340. |

|
Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. (2011) N-Terminal Acetylation Acts as an Avidity Enhancer Within an Interconnected Multiprotein Complex. Science, 334:674-678. |
|
Lee PC, Sowa SE, Gygi SP, Harper JW . (2011) Alternative Ubiquitin Activation/Conjugation Cascades Interact with N-End Rule Ubiquitin Ligases to Control Degradation of RGS Proteins. Molecular Cell 43:392-405. |
|
Tan M-K M, Lim H-J, Harper JW. (2011) SCFFBXO22 regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol., 31:3687-3699 |
|
O'Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. (2010) A Genome-wide Camptothecin Sensitivity Screen Identifies a Mammalian MMS22L-NFKBIL2 Complex Required for Genomic Stability. Mol Cell. 40:645-657. |
|
Bennett, E.J., Ruch, J., Gygi, SP, and Harper, JW. (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143:951-965. |
|
Litterman N, Ikeuchi Y, Gallardo G, O'Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A. (2011) An OBSL1-Cul7 Ubiquitin Ligase Signaling Mechanism Regulates Golgi Morphology and Dendrite Patterning. PLoS Biol. 9:e1001060. |
|
Behrends, C, Sowa ME, Gygi SP, and Harper JW. (2010) Network organization of the human autophagy system. Nature 466, 68-77. |
|
Svendsen JM, Harper JW. (2010) GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 24:521-36. |
|
Sarraf SA, Harper JW. (2010) Telomeric TuRF1 wars. Developmental Cell 18:167-168. |
|
Raman M, Harper JW. (2009) Cell biology: Stairway to the proteasome. Nature 462:585-586. |
|
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA. (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Molecular Cell. 36:39-50. |
|
Ciccia, A, Bredemeyer, AL, Sowa, ME, Terret, M-E, Jallepalli, PV, Harper, JW, and Elledge, SJ. (2009) The SIOD disorder protein SMARCAL1 is an RPA- interacting protein involved in replication fork restart. Genes and Development, 23:2415-2425. |
|
Svendsen, J., Smogorzewska, A.,Sowa, M.E., O’Connell, B.O., Gygi, S.P., Elledge, S.J., and Harper, J.W. (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell, 138:63-77. |
|
Sowa, M.E., Bennett, E.J., Gygi, S.P., and Harper, J.W. (2009) Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell, 138:389-403. |
|
Lai Xu, Mathew E. Sowa, Jing Chen, Xue Li, Steven P. Gygi, and J. Wade Harper (2009) An FTS/Hook/p107FHIP Complex Interacts with and Promotes Endosomal Clustering by the Homotypic Vacuolar Protein Sorting (HOPS) Complex. Molecular Biology of the Cell, 19:5059-71 . |
|
Schulman BA and Harper JW. Ubiquitin-like protein activation: the apex for diverse signaling pathways. Nature Reviews Molecular Cell Biology, 2009, 10:319-331. |
|
Ang XL and Harper JW. Global Protein Stability (GPS): a roadmap to cellular protein homeostasis. Nature Chemical Biology. 2009, 5:9-11 . (News and Views) |
|
Ang et al. Regulation of postsynaptic rapgap spar by polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008, 283: 29424-32. |

|
Jin et al. Differential roles for checkpoint kinases in DNA damage-dependent degradation of the Cdc25A protein phosphatase. J Biol Chem. 2008 283:19322-8. |

|
Schlabach et al., Cancer proliferation gene discovery through functional genomics. Science. 2008 319:620-4. |

|
Westbrook et al., SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008 452:370-4. |
|
Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol. 2008 15:20-2. |
|
Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007 28:739-45. |
|
Jin et al. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007 447:1135-8. |
|
Stegmeier et al.Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007 446:876-81. |
|
Hao et al. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007 26:131-43. |

|
Jin et al. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006 23:709-21. |

|
Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006 124:1133-6. |

|